Published Work
Papers
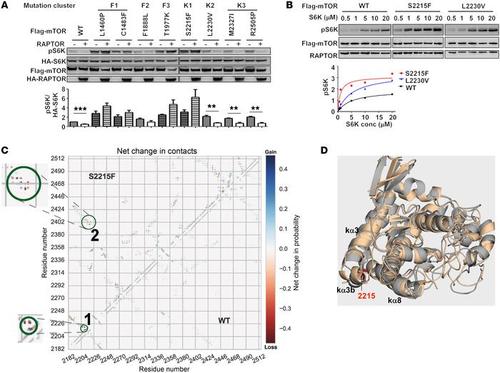 |
Mechanistically distinct cancer-associated mTOR activation clusters predict sensitivity to rapamycin Xu Jianing, Pham CG, Albanese SK, Dong Yiyu, Oyama T, Lee CH, Rodrik-Outmezguine V, Yao Z, Han S, Chen D, Parton DL, Chodera JD, Rosen N, Cheng EH, and Hsieh J. Journal of Clinical Investigation, 126:3529, 2016. DOI |
 |
An open library of human kinase domain constructs for automated bacterial expression Albanese SK Parton DL, Hanson SM, Rodriguez-Laureano L, Isik M, Behr J, Gradia S, Jeans C, Levinson NM, Seelier MA, Chodera JD. Preprint at Biorxiv, Submitted to ACS Biochemistry, DOI |
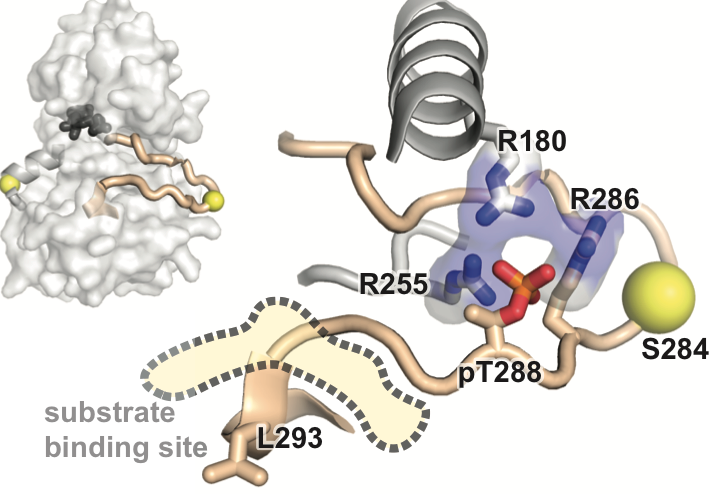 |
A dynamic mechanism for allosteric activation of Aurora kinase A by activation loop phosphorylation Ruff EF, Muretta JM, Thompson A, Lake EW, Cyphers S, Albanese SK Hanson SM, Behr JM, Thomas DD, Chodera JD, Levinson NM Preprint at Biorxiv, Submitted to eLife, DOI |
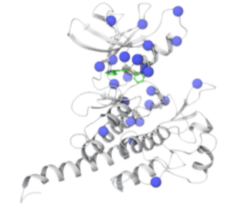 |
Predicting resistance of clinical Abl mutations to targeted kinase inhibitors using alchemical free-energy calculations Hauser K, Negron C, Albanese SK Ray S, Steinbrecher T, Abel R, Chodera JD, Wang L Preprint at Biorxiv, Submitted to Nature Comm Bio, DOI |
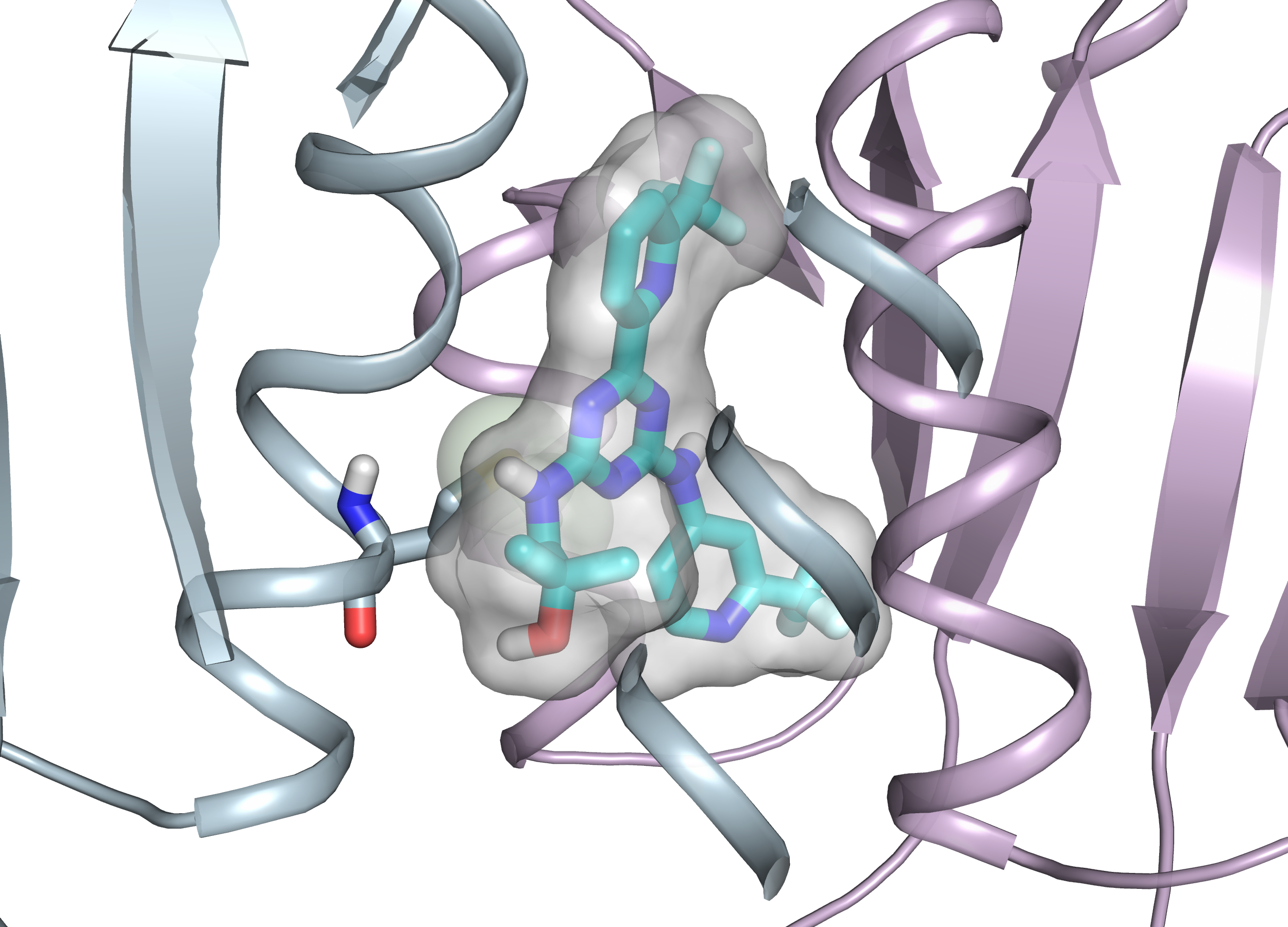 |
Acquired clinical resistance to IDH inhibition through *in trans IDH2 mutations* Intelkofer A, Shih A, Wang B, Nazir A, Rustenburg A, Albanese SK Patel M, Famulare C, Arcila M, Taylor J, Tallman M, Roshal M, Petski G, Chodera JD, Thompson C, Levine R, Stein E Submitted, DOI |
Abstracts
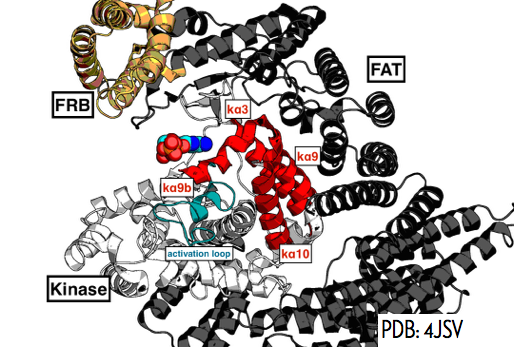 |
Simulating mTOR hyperactivating mutations to understand functionally significant structural rearrangements Steven K. Albanese, Jianing Xu, James Hsieh, John Chodera Biophysical Journal, Volume 110 , Issue 3 , 207a. DOI I submitted this abstract for the Biophysical Society Annual Meeting in Los Angeles |
Posters
 |
Simulating mTOR hyperactivating mutations to understand functionally significant structural rearrangements Steven K. Albanese, Jianing Xu, James Hsieh, John Chodera I presented this poster at the Biophysical Society Annual Meeting in Los Angeles |
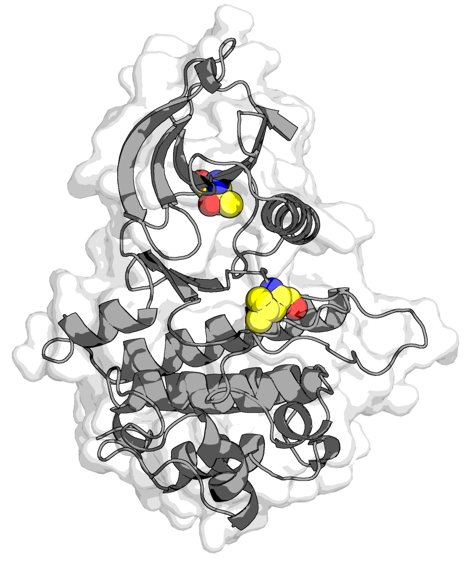 |
Rational approach to selective inhibitor design using multitarget constraints Steven K. Albanese, Patrick Grinaway, Sonya Hanson, Lucelenie Rodriguez, Zhiqiang Tan, John Chodera I presented this poster at the semi-annual Workshop on Free Energy Methods in Drug Design as well as the semi-annual Gerstner Sloan Kettering Retreat |
 |
Simulating mTOR hyperactivating mutations to understand functionally significant structural rearrangements Steven K. Albanese, Xu J, Hanson S, Fass J, Hsieh J, Chodera JD I presented this poster at the annual GTCBio Protein Kinases in Drug Discover conference |
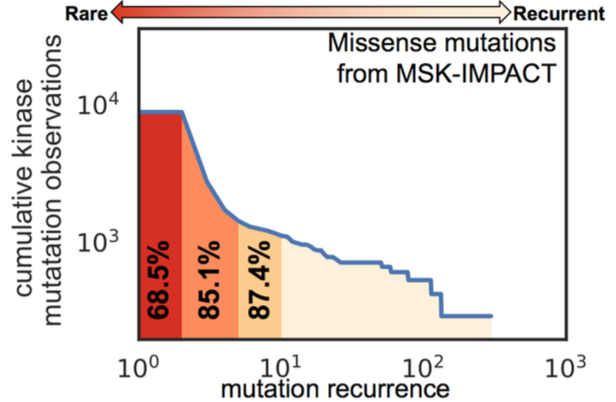 |
Using physical modeling to predict the functional impact of clinical cancer mutations in kinases on drug susceptibility Steven K. Albanese, Parton DL, Hauser K, Negron C, Hanson SM, Rodriguez-Laureano L, Isik M, Wang L, Abel R, Chodera JD A poster I put together for interviews at GSK to highlight work on bringing physical modeling and the clinic closer together |
Talks
 |
Simulating mTOR hyperactivating mutations to understand functionally significant structural rearrangements Talk I gave for ChoderaLab lab meeting, where I covered traditional MD analysis of mTOR mutants, first attempts at building an MSM for WT mTOR as well as an early attempt to improve loop modeling using alchemical softening with SAMS. |
 |
Applications of free energy methods to understand selectivity of kinase inhibitors and the function impact of mutations Talk I gave for ChoderaLab lab meeting, where I covered using FEP+ to predict selectivity of kinase inhibitors and infer the correlation of errors between different targets. I also talk about an approach using biophysical experiments and free energy calculations to understand the functional impact of clinical kinase mutations |
 |
Using physical modeling to direct biophysical experiments and predict the functional impact of clinical cancer mutations in kinases on drug susceptibility A talk I gave at the EMBO workshop on integreating genomics and biophysics in Turin, Italy in 2017. It focuses on the use of protein mutation FEP to predict resistance mutants in Abl kinase, and plans for prospective work on predicting and then measuring the calculations in Src |
 |
Using physical modeling to direct biophysical experiments and predict the functional impact of clinical cancer mutations in kinases on drug susceptibility A talk I gave at the Med Into Grad conference at Columbia on Precision Medicine in 2017. It focuses on the use of protein mutation FEP to predict resistance mutants in Abl kinase, and plans for prospective work on predicting and then measuring the calculations in Src |
 |
Is structure based drug design ready to predict selectivity A talk I gave for my yearly Graduate Student Seminar in 2017. In it, I look at the accuracy of relative and absolute free energy calculations for predicting the selectivity between two targets, as well as a brief look at using protein mutation FEP to close the thermodynamic cycle between two targets |
Thesis Proposal Exam
 |
Rational approach to selective inhibitor design using multitarget constraints Thesis Proposal for my qualifying exam |
